This preparation protocol was designed based on the protocols published in Nature Methods: “A primer to scaffolded DNA origami” (Castro et al., 2011) and in the U.S. National Library of Medicine: “DNA Origami with Double Stranded DNA as a Unified Scaffold” (Yang et al., 2012).
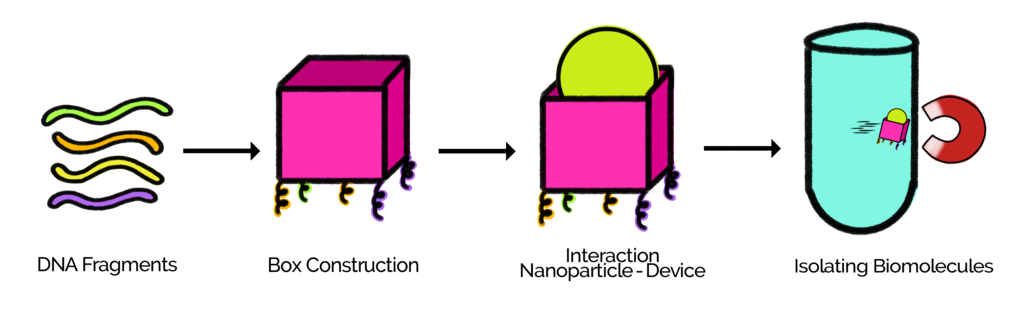
Diagram 1.Process of the construction of the box and one of its applications.
For the preparation of the device, 33 staples and 4 scaffolds were used. 33 oligonucleotides and 4 gBlocks (scaffolds) were purchased from Uniparts. Various stocks with different concentrations were prepared. Three options for the construction of the device were designed, each procedure requires specific conditions of the staples and scaffolds.
Folding reaction
A folding reaction contains scaffolds, staples, water, buffer and salt ions (Magnesium Chloride, MgCl2, was used as a salt in these experiments). The components mix due to the changes in temperature allowing the staples and scaffolds to bind together.
Due to the few available quantity of DNA, not all the desired reactions were performed. A number of folding reactions were executed varying the concentration of MgCl2 to find the optimum concentration for the design. A magnesium gradient that consisted of three different concentrations was performed. Those concentrations were 12 mM, 20 mM and 26 mM.
Standard folding reaction
A folding reaction was made with the scaffolds @1 nM and an excess of 50 staples.
Table 1. Standard Folding reaction used in the different experiments.
Thermocycler
As our scaffolds are double-stranded, they must be denatured in order to only use the desired sequence. The temperatures used in the thermocycler to denature the DNA were the following: First 90°C for 15 minutes, then it was cooled down to 25°C in less than 1 minute, after that the mixture was incubated for 2 minutes, subsequently, the mixture was heated at a temperature of 63°C, and finally it was cooled down slowly to 4°C at a rate of 20min/°C.
Gel
Electrophoresis gels were performed under the following conditions: Agarose gel was prepared at 2%. It was made with buffer TBE at 0.5X and MgCl2 at 11mM. A molecular marker (1kb Promega) was used to load the gel., as well the sample substance, that is the product of the folding reaction.
Microscope
In Air Scan. The sample (2 µL) was deposited onto a freshly cleaved mica surface and left to adsorb for 2 min. Then 50 µL of deionized H2O was added onto the mica and was blown away immediately by condensed air. The dry sample was then scanned in Nova Software in semicontact mode.
Option 1: Construction of the box in parts
For the construction of the box, four tubes were prepared (4 folding reactions), in which the staples, previously identified at a concentration of 500nM, were used. The four different scaffolds and their corresponding staples were poured in the tubes.
Therefore, each tube was composed of one scaffold, its corresponding staples, MgCl2, buffer and water.
The four folding reactions were mixed in one tube. From this tube, two aliquots were obtained. One of them was incubated in the thermocycler. Finally, both were observed in an agarose gel and characterized by AFM.
Folding reaction
Table 2. Components of the folding reaction used in option 1 "Construction of the box in parts".
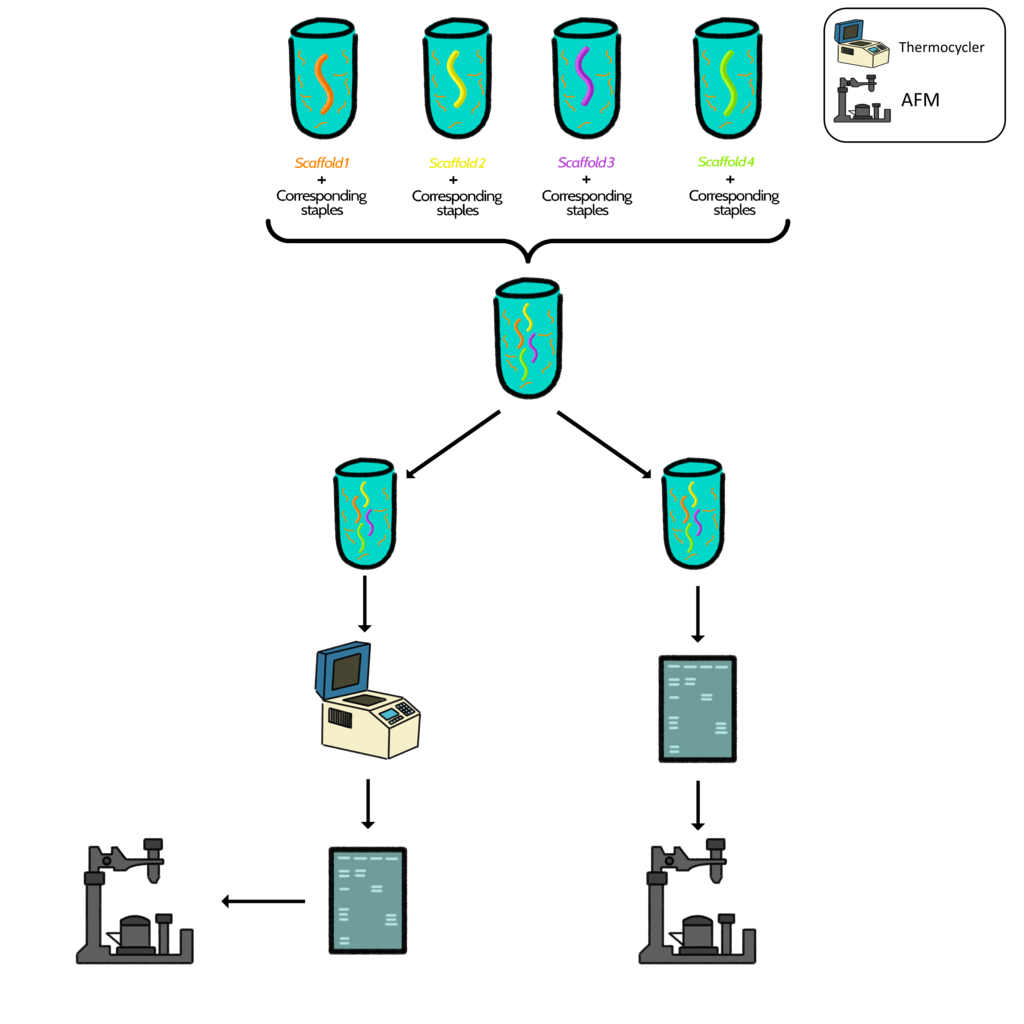
Diagram 2. Graphical representation of steps followed in option 1.
Option 2: Construction of the box ab ovo
In a tube, a folding reaction was prepared with the 4 scaffolds @1 nM, and the 33 staples @50 nM, buffer, MgCl2 and water. Then the reaction was incubated in the thermocycler, observed in an electrophoresis gel and characterized in AFM.
Folding reaction
Table 3. Components of the folding reaction used in option 2 "Construction of the box ab ovo."
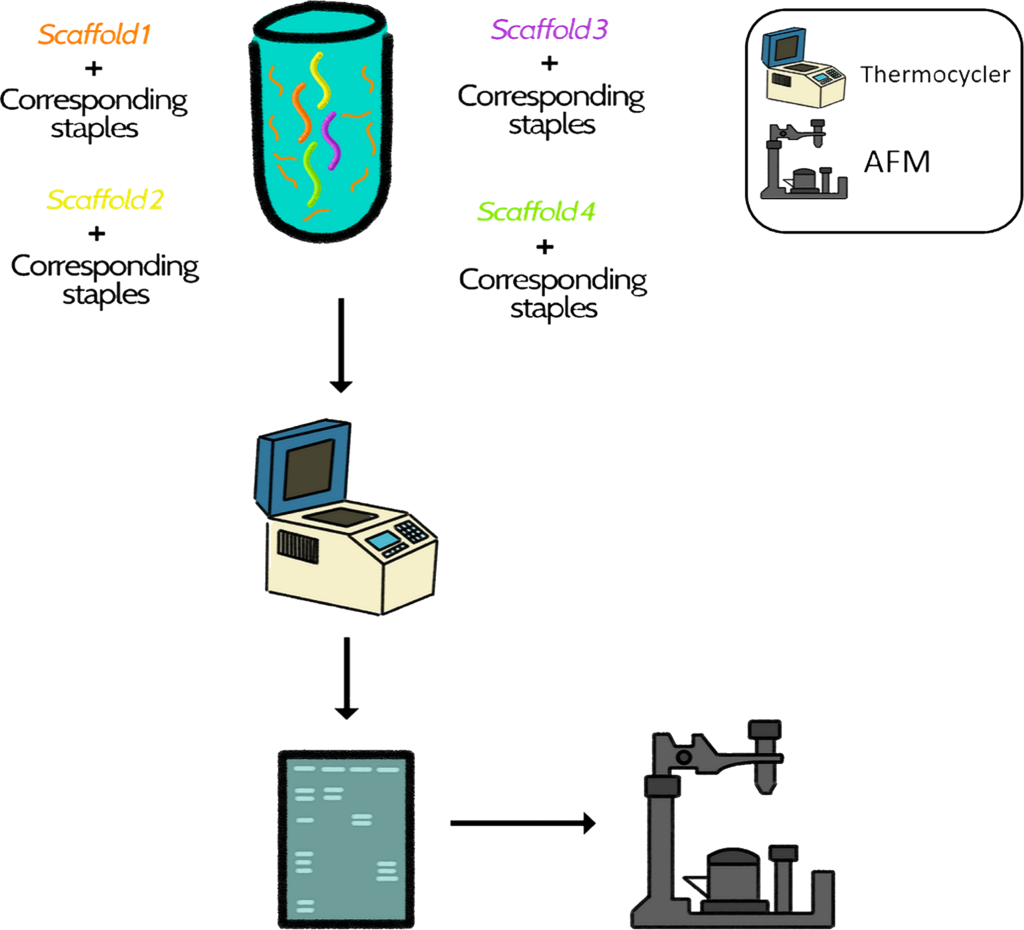
Diagram 3. Graphical representation of steps followed in option 2.
Option 3: Construction of the box step-by-step
This option consisted in joining the components of the box part by part with the following conditions:
Four folding reactions were performed separately in four tubes with one scaffold and its corresponding staples per tube. The folding reaction containing the scaffold 1 was mixed with scaffold two and incubated in the thermocycler. The new tube containing scaffold 1 and 2 was mixed with scaffold 3 and again incubated. Finally the folding reaction containing scaffolds 1,2 and 3 was combined with scaffold 4 and incubated in the thermocycler for several hours. Afterwards, the mix in the final tube was run in an agarose gel and characterized in AFM.
Folding reaction
Table 4. Components of the folding reaction used in option 3 "Construction of the box Steb-by-Step"
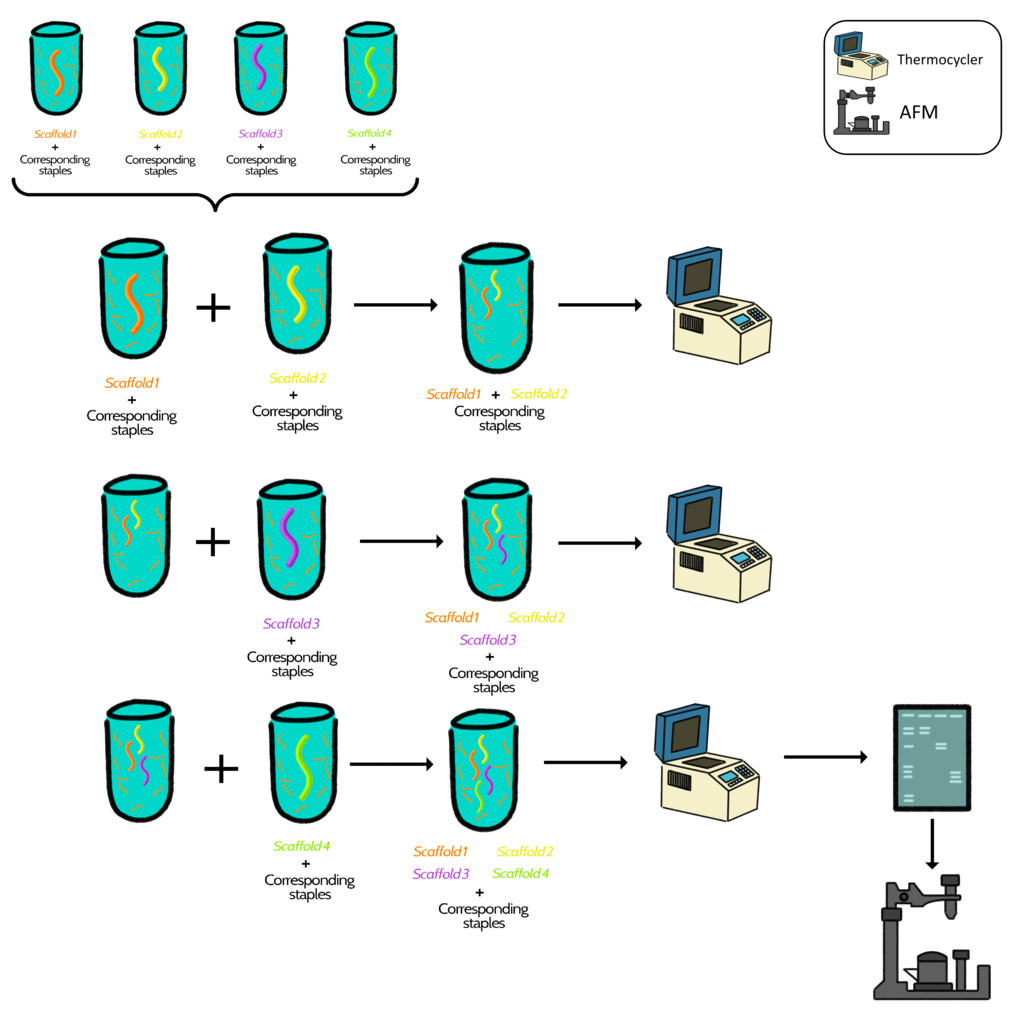
Diagram 4. Graphical representation of steps followed in option 3.
The following table shows the folding reactions performed during the experimental work made by the team.
The table presents the results in a summerized way, each column exhibits a specific condition of the results. the first column shows a code. The code permits to identify easily each folding reaction executed. It is composed of three different variables, those are temperature, concentration of MgCl2 and excess of staples. The first digit represents the final temperature used in the thermocycler, the second number stands for the concentration of MgCl2 and the third digit shows the excess of staples over scaffolds used in the reaction.
In order to use the IONPs (obtained by a donation of the Facultad de Ciencias Físico Matemáticas) in the design of the device they were functionalized with aminosilane to give them a positive charge. The following text summarizes what was done to functionalize the IONPs. (Image 1)
The IONPs were mixed with DI water and sonicated during several minutes. 25 mL of APTES diluted in water were added to the tube containing the IONPs (Image 2). After that, the tube was shaken in vortex. The solution was incubated in a laboratory oven during 3 hours at a temperature of 134°C. At last, they were washed by centrifugation at 5000 rmp/20 mins and dispersed in ethanol.

Image 1. Magnetic Iron Oxide NPs.
The image shows the magnetic property
of the NPs by stiring them
with the help of a magnet.
Image 2. Falcon tubes containing IONPs and APTES.
The uncoated IONPs from the first tube [left]
are being transferred to the tube containing APTES
second tube [right].
La Caja has magnetic properties, making it able to be controlled with a magnetic field. In order to give that property, IONPs were added to the device. These IONPs were added following this criteria: a mix of DNA and IONPs is made. 5 µL of each substance is poured into an Eppendorf tube and resuspended in order to mix them properly. It is important to mention that the IONPs were centrifuged to separate them by size; the supernatant was taken because this contained the smaller IONPs needed to the characteristics of the device. The mix made was incubated during several minutes at room temperature.
After the construction and functionalization of the device it is necessary to verify the arrangement of the box using different methods as microscopes and spectrometers. The AFM (Image 3) and TEM (Image 4) were used, as well as the Fourier Transform Infrared Spectroscopy (FTIR). The following text describe what was made in each one of them. The AFM was used in order to characterize the device, the technique used was in air scan. The TEM and FTIR were used to prove the functionalization of the device. TEM showed different images of the IONPs before and after the functionalization. As a second source to ascertain the fuctionalization of the IONPs the FTIR was used obtaining graphical representations of the coated and uncoated IONPs. In addition to the FTIR, the Zeta Potential was measured with a Zeta Sizer.

Image 3. Atomic Force
Microscope (AFM).

Image 4. Transmission Electron
Microscope (TEM).
This section presents the protocol that was followed by the team to perform the different experiments. It was designed based on the protocols published in Nature Methods: “A primer to scaffolded DNA origami” (Castro et al., 2011) and in the U.S. National Library of Medicine: “DNA Origami with Double Stranded DNA as a Unified Scaffold” (Yang et al., 2012).
Castro, C. E., Kilchherr, F., Kim, D. N., Shiao, E. L., Wauer, T., Wortmann, P., ... & Dietz, H. (2011). A primer to scaffolded DNA origami. Nature methods, 8 (3), 221-229.
Nakagawa, T., Hashimoto, R., Maruyama, K., Tanaka, T., Takeyama, H., & Matsunaga, T. (2006). Capture and release of DNA using aminosilane‐modified bacterial magnetic particles for automated detection system of single nucleotide polymorphisms. Biotechnology and bioengineering, 94 (5), 862-868.
Nakagawa, T., Tanaka, T., Niwa, D., Osaka, T., Takeyama, H., & Matsunaga, T. (2005). Fabrication of amino silane-coated microchip for DNA extraction from whole blood. Journal of biotechnology, 116 (2), 105-111.
Ota, H., Lim, T. K., Tanaka, T., Yoshino, T., Harada, M., & Matsunaga, T. (2006). Automated DNA extraction from genetically modified maize using aminosilane-modified bacterial magnetic particles. Journal of biotechnology, 125 (3), 361-368.
Yang, Y., Han, D., Nangreave, J., Liu, Y., & Yan, H. (2012). DNA origami with double-stranded DNA as a unified scaffold. ACS nano, 6 (9), 8209-8215.